Evaluation of genetic variation in longan (Dimocarpus longan Lour) by high annealing temperature random amplified polymorphic DNA (HAT−RAPD)
DOI:
https://doi.org/10.55674/cs.v15i2.250017Keywords:
Dimocarpus longan Lour, High annealing temperature RAPD, Genetic variation, Molecular markerAbstract
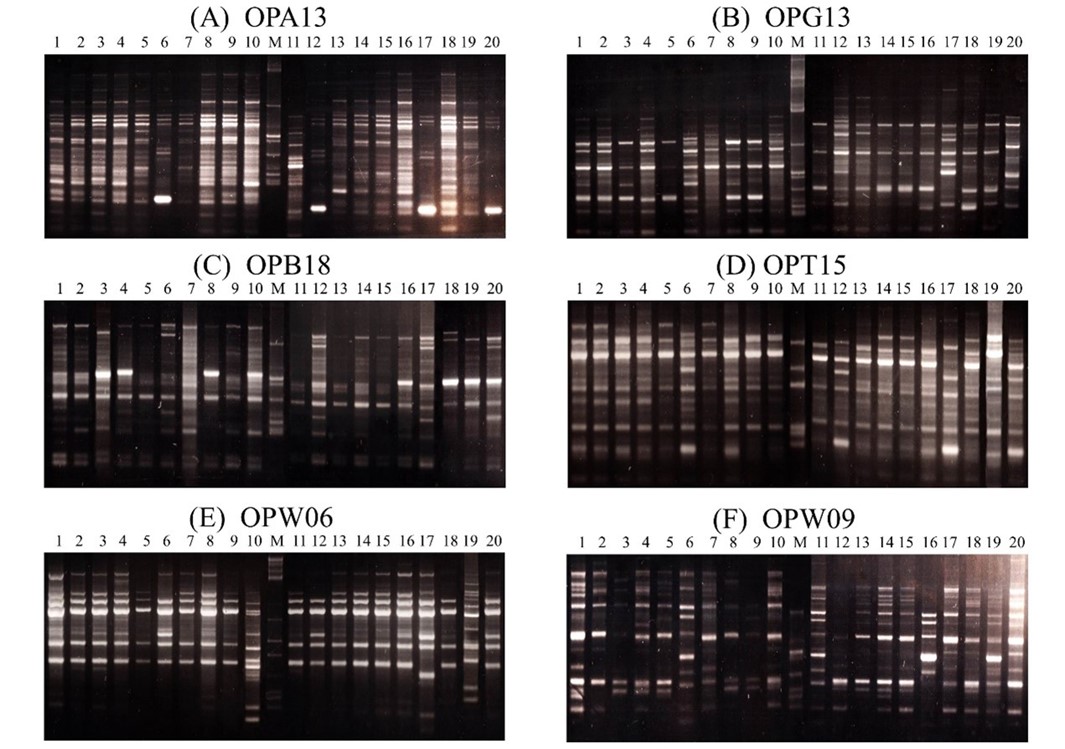
Genetic relationships among 20 longan varieties (Dimocarpus longan Lour.) collected from Chiang Rai and Lampang province; Thailand, Vietnam and China were investigated using high annealing temperature random amplified polymorphic DNA (HAT−RAPD) technique. Ten primers, OPA 13, OPAK10, OPB 18, OPG 13, OPW 06, OPW 09, OPX 01, OPX 15, OPZ 01 and OPAK 10, produced a total of 305 RAPD markers. All primers generated a high degree of polymorphism, ranging in molecular weight from 100 to 2,500 bp. Hierarchical cluster analysis, based on unweighted neighbor−joining method classified the 20 varieties into 3 major groups. The first two groups could be divided into 2 subgroups and the third group contained only one subgroup. Our data revealed that members in the first group are Daw types.
The Narapirom and Petchsakorn varieties exhibited close relatedness, suggesting that they are originated from the same variety. In addition, this technique also detected variation among longan varieties due to selective breeding.
References
D.S. Mishra, B. Chakraborty, H. Rymbai, N. Deshmukh, A.K. Jha, G.F. War, D. Paul, R.S. Patel, L.K. Mishra, D. Roy, P. Lyngdoh, Breeding of Underutilized Fruit Crops : Longan (Dimocarpus longan Lour). aya Pub, House, Delhi, 2018.
J.G.K. Williams, A.R. Kubelik, K.J. Livak, J.A. Rafalski, S.V. Tingey, DNA polymorphisms amplified by arbitrary primers are useful as genetic markers, Nucleic Acids Res. 18 (19) (1990) 6531 − 6535.
J. Welsh, M. McClelland, Fingerprinting genomes using PCR with arbitrary primers, Nucleic Acids Res. 18 (24) (1990) 7213 − 7218.
T. Arumugam, G. Jayapriya, T. Sekar, Molecular fingerprinting of the Indian medicinal plant Strychnos minor Dennst, Biotechnol. Rep. 21 (2019) e00318.
Y. Duran, W. Rohde, A. Kullaya, P. Goikoetxea, E. Ritter, Molecular analysis of East African tall coconut genotypes by DNA marker technology, J. Genet. Breed. 51 (4) (1997) 279 − 288.
V.R. Hinge, I.M. Shaikh, R.L. Chavhan, A.S. Deshmukh, R.M. Shelake, S.A. Ghuge, A.M. Dethe, P. Suprasanna, U.S. Kadam, Assessment of genetic diversity and volatile content of commercially grown banana (Musa spp.) cultivars, Sci. Rep. 12 (2022) 7979.
P.J. Elisiario, M. Justo, J.M. Leitao, Identification of mandarin hybrids by isozyme and RAPD analysis, Sci. Hort. 81 (3) (1999) 287 − 299.
M. Pharmawati, I.J. Macfarlane, The genetic relationships of grevillea hybrids determined by RAPD marker, HAYATI J. Biosci. 20 (4) (2013) 196 − 200.
A. Kaga, N. Tomooka, Y. Egawa, K. Hosaka, O. Kamijima, Species relationships in the subgenus Ceratotropis (genus Vigna) as revealed by RAPD analysis, Euphytica. 88 (1996) 17 − 24.
L.F. Cabrita, U. Aksoy, S. Hepaksoy, J.M. Leitao, Suitability of isozyme, RAPD and AFLP markers to assess genetic differences and relatedness among fig (Ficus carica L.) clones, Sci. Hort. 87 (4) (2001) 261 − 273.
P. Sharma, A.K. Nath, S.R. Dhiman, S. Dogra, V. Sharma, Characterization of carnation (Dianthus caryophyllus L.) genotypes and gamma irradiated mutants using RAPD, ISSR and SSR markers, S.Afr.J.Bot. 148 (2022) 67 − 77.
D. Dhakshanamoorthy,R. Selvaraj, A. Chidambaramb, Utility of RAPD marker for genetic diversity analysis in gammarays and ethyl methane sulphonate (EMS)-treated Jatropha curcas plants, Comptes Rendus Biologies, 338 (2014) 75 – 82.
R. Aslam, T.M. Bhat, S. Choudhary, M.Y.K. Ansari, D. Shahwar, Estimation of genetic variability, mutagenic effectiveness and efficiency in M2 flower mutant lines of Capsicum annuum L. treated with caffeine and their analysis through RAPD markers, J. King Saud Univ. Sci. 29 (3) (2017) 274 − 283.
D. Wahyudi, L. Hapsari, S. Sundari, RAPD analysis for Geneti variability detection of mutant soybean (Glycine max (L.) Merr), J. Tropical Biodiversity Biotechnology. 05(1) (2020) 68 – 77.
S. Anuntalabhochai, R. Chundet, J. Chiangda, P. Apavatjrut, Genetic diversity within Lychee (Litchi chinensis Sonn.) based on RAPD analysis, Acta Hortic. 575 (2002) 253 − 259.
T.W. Chen, C.C. NG, C.Y. Wang, Y.T. Shyu, Molecular identification and analysis of Psidium guajava L. from indigenous tribes of Taiwan, J. Food Drug Anal. 15 (1) (2007) 82 − 88.
M.N.R. Baig, S. Grewal, S. Dhillon, Molecular characterization and genetic diversity analysis of citrus cultivars by RAPD markers, Turk J. Agric For. 33 (4) (2009) 375 − 384.
U. Erturk, M.E. Akcay, Genetic variability in accessions of “Amasya” apple cultivar using RAPD markers, Not. Bot. Hort. Agrobot. Cluj. 38 (3) (2010) 239 − 245.
F.P. Nicese, J.I. Hotrmaza, G.H. McGranahan, Molecular characterization and genetic relatedness among walnut (Juglans regia L.) genotypes based on RAPD markers, Euphytica. 101 (1998) 199 − 206.
G.A. Penner, A. Bush, R. Wise, W. Kim, L. Domier, K. Kash, A. Laroche, G. Scoles, S.J. Molnar, G. Fedak, Reproducibility of random amplified polymorphic DNA (RAPD) analysis among laboratories, PCR Methods Appls. 2 (4) (1993) 341 − 345.
Y. Shoyama, F. Kawachi, H. Tanaka, R. Nakai, T. Shibata, K. Nishi, Genetic and alkaloid analysis of Papaver species and their F1 hybrid by RAPD, HPLC and Elisa, Forensic Sci. Int. 91 (3) (1998) 207 − 217.
T. Shimada, H. Hayama, T. Haji, M. Yamaguchi, M. Yoshida, Genetic diversity of plums characterized by random amplified polymorphic DNA (RAPD) analysis, Euphytica. 109 (1999) 143 − 147.
F. Atienzar, A. Evanden, A. Jha, D. Savva, M. Depledge, Optimized RAPD analysis generates high quality genomic DNA profiles at high annealing temperature, BioTechniques. 28 (1) (2000) 52 − 54
S. Anuntalabhochai, W. Phromthep, S. Sitthiphrom, R. Chundet, R.W. Cutler, phylogenetic diversity of ficus species using HAT-RAPD markers as a measure of genomic polymorphism, Open Agric. 2 (2008) 62 – 67.
J. Jiemjuejun, S. Damrianant, T. Thanananta, N. Thanananta, Genetic Relationship assessment and identification of Orchids in the Geenus Eria Using HAT-RAPD Markers, Sci. & Tech. Asia. 22 (2017) 19 – 26.
T. Meesangiem, S. Damrianant, T. Thanananta, N. Thanananta, Genetic Relationship among Paphiopedilum subgenus Brachypetalum section Brachypetalum using HAT-RAPD Markers, Thai J. of Science and Technology, 1 (2018) 99 – 105.
J.J. Doyle, J.L. Doyle, A rapid DNA isolation procedure for small quantities of fresh leaf tissue, Phytochem Bull. 19 (1) (1987) 11 − 15.
P. Kunasol, Increasing period of longan production, House Agric Magazine. 21 (1995) 106 − 112.
N.P. Songkhla, Visiting longan orchard in Vietnam, House Agric. Magazine. 24 (2000) 83 − 88.
I.A. Arif, M.A. Bakir, H.A. Khan, A.H. Al Farhan, A.A Al Homaidan, A.H. Bahkali, M. Al Sadoon, M. Shobrak, Application of RAPD for molecular characterization of plant species of medicinal value from an arid environment, Genet Mol Res. 9 (4) (2010) 2191 – 2198.
Y. Fei, W. Tang, J. Shen, Z. Tianjing, Q. Rui, B. Xiao, C. Zhou, Z. Liu, Y.T. Anna, Application of random amplified polymorphic DNA (RAPD) markers to identify Taxus chinensis var. mairei cultivars associated with parthenogenesis, Afr. J. Biotechnol., 13 (4) (2014) 2385 – 2393.
P. Butboonchoo, C. Wongsawad, Occurrence and HAT-RAPD analysis of gastrointestinal helminths in domestic chickens (Gallus gallus domesticus) in Phayao province, northern Thailand, Saudi J. Biol. Sci., 24 (2017) 30 – 35.
E. Rybska, A. Pacak, Z. Szweykowska-Kulinska, A. Lesicki, RAPD Markers as a tool for analysis of relationships among selected species of Lymnaeidae (Gastropoda Pulmonata), Filia Malacol., 16(1) (2008) 39 – 51.
Downloads
Published
How to Cite
Issue
Section
Categories
License
Copyright (c) 2023 Creative Science

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.








