Serum glycobiomarker mining suggested the improvement of cholangiocarcinoma detection using combined CA125 and CA242
Main Article Content
Abstract
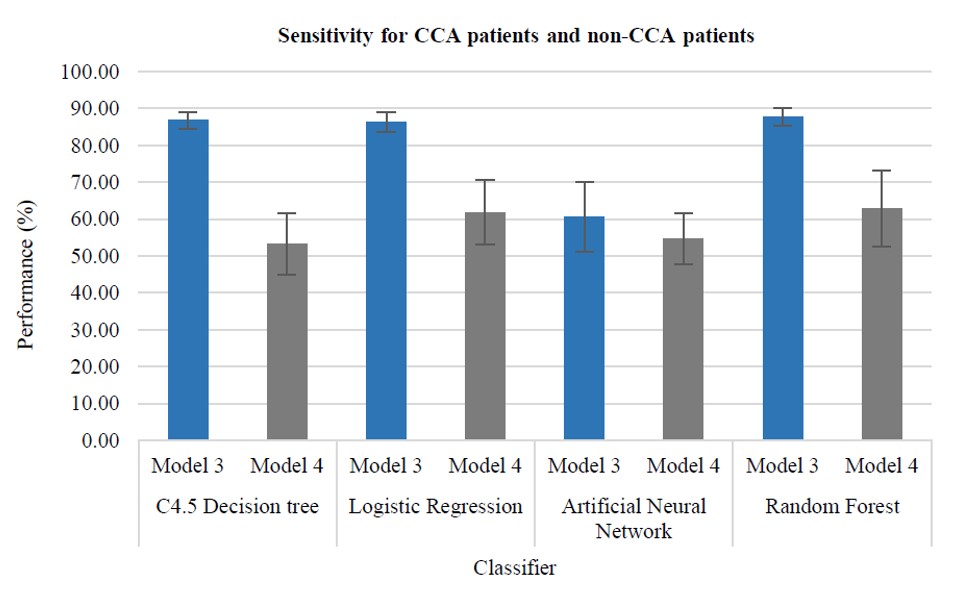
Cholangiocarcinoma (CCA) is a malignant neoplasm originating from biliary epithelial cells. During the early stage, the patients do not show any symptoms, leading to wide and extensive spread of this disease. Nowadays, there has not been a single serum tumor marker which can be used for effective screening of the disease or classification of the patients. This study therefore aims to determine an appropriate serum marker for screening of the patients with early staged CCA by using a technique of data mining. Beginning with the C4.5 Decision tree and Logistic Regression for selection of serum markers for effective screening of the patients with CCA, the selected markers were then used for classification of the patients with CCA from non-CCA patients, and CCA from Benign Biliary Disease (BBD) by C4.5 Decision tree, Logistic Regression, Random Forest, and Artificial Neural Network. In this work, seven serum tumor markers were used, including Carbohydrate Antigen 125 (CA125), Carbohydrate Antigens 19-9 (CA19-9), Carbohydrate Antigen 242 (CA242), Carbohydrate Antigen 50 (CA50), Carbohydrate Antigen 72-4 (CA72-4), Carcinoembryonic Antigen (CEA), Cy-tokeratin-19Fragment (CYFRA 21-1). The model was used to classify the CCA and non-CCA patients and it was discovered that the serum tumor markers which could most efficiently classify the CCA patients from the non-CCA patients were the combination of CA125 and CA242 suggested by the Logistic Regression with C4.5 Decision tree as the classifier, yielding the best performance, with Sensitivity and Specificity being 75.88 % and 86.82%, respectively. In contrast, the classification of CCA patients from BBD patients was best performed by the serum tumor markers CA125 and CA72-4 suggested by C4.5 Decision tree with Logistic Regression or Random Forest as the classifier.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
References
Goral V. Cholangiocarcinoma: new insights. Asian Pac J Cancer Prev. 2017;18(6):1469-73.
Chaiteerakij R, Pan-ngum W, Poovorawan K, Soonthornworasiri N, Treeprasertsuk S, Phaosawasdi K. Characteristics and outcomes of cholangiocarcinoma by region in Thailand: a nationwide study. World J Gastroenterol. 2017;23(39):7160-7.
Khan SA, Tavolari S, Brandi G. Cholangiocarcinoma: epidemiology and risk factors. Liver Int. 2019;39(S1):19-31.
Seeherunwong A, Chaiear N, Khuntikeo N, Ekpanyaskul C. The proportion of occupationally related cholangiocarcinoma: a tertiary hospital study in Northeastern Thailand. Cancers (Basel). 2022;14(10):2386.
Uenishi T, Yamazaki O, Tanaka H, Takemura S, Yamamoto T, Tanaka S, et al. Serum cytokeratin 19 fragment (CYFRA21-1) as a prognostic factor in intrahepatic cholangiocarcinoma. Ann Surg Oncol. 2008;15(2):583-9.
Zhang Y, Yang J, Li H, Wu Y, Zhang H, Chen W. Tumor markers CA19-9, CA242 and CEA in the diagnosis of pancreatic cancer: a meta-analysis. Int J Clin Exp Med. 2015;8(7):11683-91
Qiu Y, He J, Chen X, Huang P, Hu K, Yan H. The diagnostic value of five serum tumor markers for patients with cholangiocarcinoma. Clin Chim Acta. 2018;480:186-92.
Luang S, Teeravirote K, Saentaweesuk W, Ma-In P, Silsirivanit A. Carbohydrate antigen 50: values for diagnosis and prognostic prediction of intrahepatic cholangiocarcinoma. Medicina. 2020;56(11):616.
Wongkham S, Silsirivanit A. State of serum markers for detection of cholangiocarcinoma. Asian Pac J Cancer Prev. 2012;13(Suppl):17-27.
Pattanapairoj S, Silsirivanit A, Muisuk K, Seubwai W, Cha’on U, Vaeteewoottacharn K, et al. Improve discrimination power of serum markers for diagnosis of cholangiocarcinoma using data mining-based approach. Clin Biochem. 2015;48(10-11):668-73.
Song HJ, Yang ES, Kim JD, Park CY, Kyung MS, Kim YS. Best serum biomarker combination for ovarian cancer classification. Biomed Eng Online. 2018;17(S2):152.
Kimawaha P, Jusakul A, Junsawang P, Thanan R, Titapun A, Khuntikeo N, et al. Establishment of a potential serum biomarker panel for the diagnosis and prognosis of cholangiocarcinoma using decision tree algorithms. Diagnostics (Basel). 2021;11(4):589.
Rustam Z, Zhafarina F, Saragih GS, Hartini S. Pancreatic cancer classification using logistic regression and random forest. IAES Int J Artif Intell. 2021;10(2):476-81.
Mahesh TR, Vinoth Kumar V, Dhilip Kumar V, Geman O, Margala M, Guduri M. The stratified K-folds cross-validation and class-balancing methods with high-performance ensemble classifiers for breast cancer classification. Healthcare Analytics. 2023;4:100247.
Botlagunta M, Botlagunta MD, Myneni MB, Lakshmi D, Nayyar A, Gullapalli JS, et al. Classification and diagnostic prediction of breast cancer metastasis on clinical data using machine learning algorithms. Sci Rep. 2023;13(1):485.
Chandrashekar K, Setlur AS, Sabhapathi CA, Raiker SS, Singh S, Niranjan V. Decision support system and web-application using supervised machine learning algorithms for easy cancer classifications. Cancer Inform. 2023;22:1-18.
Bishop CM. Pattern recognition and machine learning. 2nd ed. New York: Springer; 2006.
Smith DR. Top-down synthesis of divide-and-conquer algorithms. Artif Intell. 1985;27(1):43-96.
Salzberg SL. C4.5: programs for machine learning by J. Ross Quinlan. Morgan Kaufmann Publishers, Inc., 1993. Mach Learn. 1994;16(3):235-40.
Cramer JS. The origins of logistic regression. Tinbergen Institute Working Paper No. 2002-119/4. Amsterdam: Tinbergen Institute; 2002.
Lu K. Logistic regression in biomedical study. 2022 International Conference on Biotechnology, Life Science and Medical Engineering (BLSME 2022); 2022 Jan 22-23; Jeju, South Korea. p. 589-95.
Ahmad I, Basheri M, Iqbal MJ, Rahim A. Performance comparison of support vector machine, random forest, and extreme learning machine for intrusion detection. IEEE Access. 2018;6:33789-95.
Wu Z, Lin W, Zhang Z, Wen A, Lin L. An ensemble random forest algorithm for insurance big data analysis. 2017 IEEE International Conference on Computational Science and Engineering (CSE) and IEEE International Conference on Embedded and Ubiquitous Computing (EUC); 2017 Jul 21-24; Guangzhou, China. New York: IEEE; 2017. p. 531-6.
Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323(6088):533-6.
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. ACM SIGKDD Explorations Newsletter. 2009;11(1):10-8.
Rim P, Liu E. Optimizing the C4.5 decision tree algorithm using MSD-Splitting. IJACSA. 2020;11(10):41-7.