White Blood Cell Classification Using SMOTE-SVM Method with Hybrid Feature Extraction and Image Segmentation Using Gaussian Mixture Model
Main Article Content
Abstract
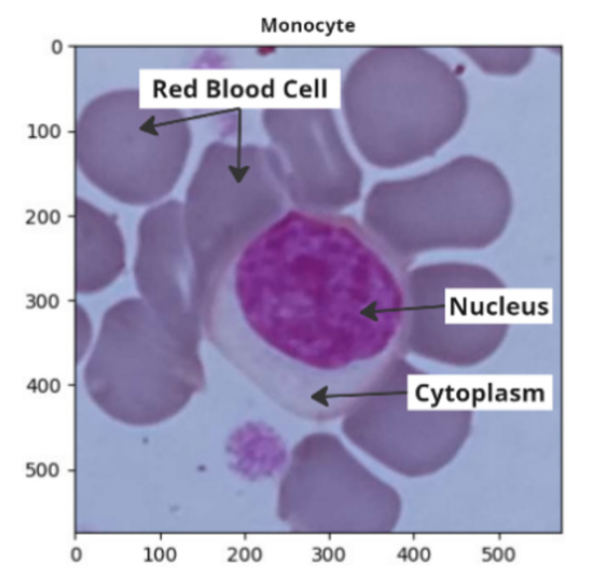
White blood cells are crucial to the immune system. The irregular structure of white blood cells, along with the fact that each type has its unique structure, makes manual identification challenging. Manual identification is prone to errors due to medical personnel's subjectivity and fatigue from time and effort demands. A fast and accurate method for classifying white blood cells is needed, but challenges remain regarding the quality and quantity of samples for each cell type. This study proposes the use of SMOTE and SVMSMOTE to address the issue of data imbalance, as well as a combination of shape features (size, circularity, convexity, solidity) and convolutional autoencoder (CAE) for feature extraction, along with a Gaussian mixture model for nucleus segmentation. The study finds that, without using SMOTE or SVMSMOTE for data balancing, the proposed features are already sufficient to represent each cell type except eosinophils, achieving an accuracy of 92.4%, precision of 91.9%, recall of 92.3%, F1-Score of 92%, MCC of 0.862, and CEN of 0.1376 using a polynomial kernel. The worst results were obtained with the sigmoid kernel. The combined feature extraction (shape and CAE) outperformed individual methods. Shape alone achieved 86.8% accuracy, CAE alone 87.8%. Recall for eosinophil cells improved using SMOTE and SVMSMOTE.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
References
J. Goretzko et al., “P-selectin-dependent leukocyte adhesion is governed by endolysosomal twopore channel 2,” Cell Rep, vol. 42, no. 12, p. 113501, 2023.
M. Zhu, W. Chen, Y. Sun and Z. Li, “ImprovedU-net-based leukocyte segmentation method,” J Biomed Opt, vol. 28, no. 04, Apr. 2023.
B. S. S. Rao and B. S. Rao, “An Effective WBC Segmentation and Classification Using MobilenetV3 ShufflenetV2 Based Deep Learning Framework,” IEEE Access, vol. 11, pp. 27739–27748, 2023.
O. Katar and O. Yildirim, “An Explainable Vision Transformer Model Based White Blood Cells Classification and Localization,” Diagnostics, vol. 13, no. 14, Jul. 2023.
Z. M. Kouzehkanan et al., “A large dataset of white blood cells containing cell locations and types, along with segmented nuclei and cytoplasm,” Sci Rep, vol. 12, no. 1, Dec. 2022.
H. Chen et al., “Accurate classification of white blood cells by coupling pre-trained ResNet and DenseNet with SCAM mechanism,” BMC Bioinformatics, vol. 23, no. 1, Dec. 2022.
S. Rashid, M. Raza, M. Sharif, F. Azam, S. Kadry and J. Kim, “White blood cell image analysis for infection detection based on virtual hexagonal trellis (VHT) by using deep learning,” Sci Rep, vol. 13, no. 1, Dec. 2023.
L. Zucchini et al., “Characterization of a Novel Approach for Neonatal Hematocrit Screening Based on Penetration Velocity in Lateral Flow Test Strip,” Sensors, vol. 23, no. 5, Mar. 2023.
I. Lin, O. Loyola-Gonz´alez, R. Monroy and M. A. Medina-P´erez, “A review of fuzzy and pattern-based approaches for class imbalance problems,” Appl. Sci., vol. 11, no. 14:6310, 2021.
L. Wang, M. Han, X. Li, N. Zhang and H. Cheng, “Review of Classification Methods on Unbalanced Data Sets,” IEEE Access, vol. 9, pp. 64606–64628, 2021.
M. Koziarski, “Potential Anchoring for imbalanced data classification,” Pattern Recognition, vol. 120, Dec. 2021.
S. Khan, M. Sajjad, T. Hussain, A. Ullah and A. S. Imran, “A review on traditional machine learning and deep learning models for WBCs classification in blood smear images,” IEEE Access, vol. 9, pp. 10657–10673, 2021.
S. Tavakoli, A. Ghaffari, Z. M. Kouzehkanan and R. Hosseini, “New segmentation and feature extraction algorithm for classification of white blood cells in peripheral smear images,” Sci Rep, vol. 11, no. 1, Dec. 2021.
L. S. Lin, C. H. Kao, Y. J. Li, H. H. Chen and H. Y. Chen, “Improved support vector machine classification for imbalanced medical datasets by novel hybrid sampling combining modified mega-trend diffusion and bagging extreme learning machine model,” Mathematical Biosciences and Engineering, vol. 20, no. 10, pp. 17672–17701, 2023.
S. Devella, Y. Yohannes and C. Adi Putra, “Penggunaan Fitur Saliency-SURF Untuk Klasifikasi Citra Sel Darah Putih Dengan Metode SVM,” vol. 8, no. 4, 2021.
Y. Yohannes, S. Devella and W. Hadisaputra, “Pemanfaatan Scale Invariant Feature Transform Berbasis Saliency untuk Klasifikasi Sel Darah Putih,” Jurnal Teknik Informatika dan Sistem Informasi, vol. 7, no. 2, Aug. 2021.
F. Riaz et al., “Gaussian Mixture Model Based Probabilistic Modeling of Images for Medical Image Segmentation,” IEEE Access, vol. 8, pp. 16846–16856, 2020.
P. Wang, E. Fan and P. Wang, “Comparative analysis of image classification algorithms based on traditional machine learning and deep learning,” Pattern Recognit Lett, vol. 141, pp. 61–67, Jan. 2021.
M. M. Paco Ramos, V. M. Paco Ramos, A. L. Fabian, and E. F. Osco Mamani, “A Feature Extraction Method Based on Convolutional Autoencoder for Plant Leaves Classification,” in Communications in Computer and Information Science, Springer, pp. 143–154, 2019.
H. Mahmood, T. Mehmood and L. A. Al-Essa, “Optimizing Clustering Algorithms for Anti-Microbial Evaluation Data: A Majority Score Based Evaluation of K-Means, Gaussian Mixture Model, and Multivariate T-Distribution Mixtures,” IEEE Access, vol. 11, pp. 79793–79800, 2023.
K. Al-Dulaimi, J. Banks, K. Nguyen, A. AlSabaawi, I. Tomeo-Reyes and V. Chandran, “Segmentation of White Blood Cell, Nucleus and Cytoplasm in Digital Haematology Microscope Images: A Review-Challenges, Current and Future Potential Techniques,” IEEE Rev Biomed Eng, vol. 14, pp. 290–306, 2021.
W. F. Lamberti, “Blood cell classification using interpretable shape features: A Comparative Study of SVM models and CNN-Based approaches,” Computer Methods and Programs in Biomedicine Update, vol. 1, Jan. 2021.
N. Louanjli et al., “Infiltration of Leukocytes into the Human Ejaculate and its Association with Semen Quality and Oxidative Stress with Sperm Function, and Leukocytospermia Management,” 2021.
S. Mahajan, A. Raina, X.-Z. Gao and A. K. Pandit, “Plant Recognition Using Morphological Feature Extraction and Transfer Learning over SVM and AdaBoost,” Symmetry, vol. 13, no. 2:356, 2021.
M. Irfan, Z. Jiangbin, M. Iqbal, Z. Masood and M. H. Arif, “Knowledge extraction and retention based continual learning by using convolutional autoencoder-based learning classifier system,” Inf Sci (N Y), vol. 591, pp. 287–305, 2022.
E. Pintelas, I. E. Livieris and P. E. Pintelas, “A convolutional autoencoder topology for classification in high dimensional noisy image datasets,” Sensors, vol. 21, no. 22, Nov. 2021.
J. Sonawane, M. Patil and G. Birajdar, “A novel feature extraction and mapping using convolutional autoencoder for enhancement of Under water image/video,” ITM Web of Conferences, vol. 44, p. 03066, 2022.
M. Asrol, P. Papilo, and F. E. Gunawan, “Support Vector Machine with K-fold Validation to Improve the Industry’s Sustainability Performance Classification,” in Procedia Computer Science, Elsevier B.V., pp. 854–862, 2021.
S. Wang, Y. Dai, J. Shen and J. Xuan, “Research on expansion and classification of imbalanced data based on SMOTE algorithm,” Sci Rep, vol. 11, no. 1, Dec. 2021.
J. H. Joloudari, A. Marefat, M. A. Nematollahi, S. S. Oyelere and S. Hussain, “Effective Class-Imbalance Learning Based on SMOTE and Convolutional Neural Networks,” Applied Sciences (Switzerland), vol. 13, no. 6, Mar. 2023.
A. Kim and I. Jung, “Optimal selection of resampling methods for imbalanced data with high complexity,” PLoS One, vol. 18, Jul. 2023.
M. Khushi et al., “A Comparative Performance Analysis of Data Resampling Methods on Imbalance Medical Data,” IEEE Access, vol. 9, pp. 109960–109975, 2021.
D. Mustafa Abdullah and A. Mohsin Abdulazeez, “Machine Learning Applications based on SVM Classification A Review,” Qubahan Academic Journal, vol. 1, Nov. 2021.
A. C. Kemila, W. Fawwaz and A. Maki, “Parameter Optimization of Support Vector Machine using River Formation Dynamic on Brain Tumor Classification,” Open Access Journal, vol. 5, no. 3, pp. 177–184, 2023.
T. Ke et al., “A general maximal margin hypersphere SVM for multi-class classification,” Expert Syst Appl, vol. 237, p. 121647, 2024.
R. Yuranda, T. Sutabri and D. Wahyuningsih, “Machine Learning Approach in Evaluating News Labels Based on Titles: Online Media Case Study,” Jurnal Sisfokom (Sistem Informasi dan Komputer), vol. 12, no. 3, pp. 434–439, Nov. 2023.
S. Pertiwi, D. Handoko Wibowo and S. Widodo, “Deep Learning Model for Identification of Diseases on Strawberry (Fragaria sp.) Plants,” vol. 13, no. 4, 2023.
D. Chicco and G. Jurman, “The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation,” BMC Genomics, vol. 21, no. 1, Jan. 2020.
K. Y. Foo et al., “Multi-class classification of breast tissue using optical coherence tomography and attenuation imaging combined via deep learning,” Biomed Opt Express, vol. 13, no. 6, pp. 3380–3400, 2022.
B. Krawczyk, C. Bellinger, R. Corizzo and N. Japkowicz, “Undersampling with Support Vectors for Multi-Class Imbalanced Data Classification,” in Proceedings of the International Joint Conference on Neural Networks, Institute of Electrical and Electronics Engineers Inc., Jul. 2021.