Deep Learning-based Ensemble Approach for Conventional Pap Smear Image Classification
Main Article Content
Abstract
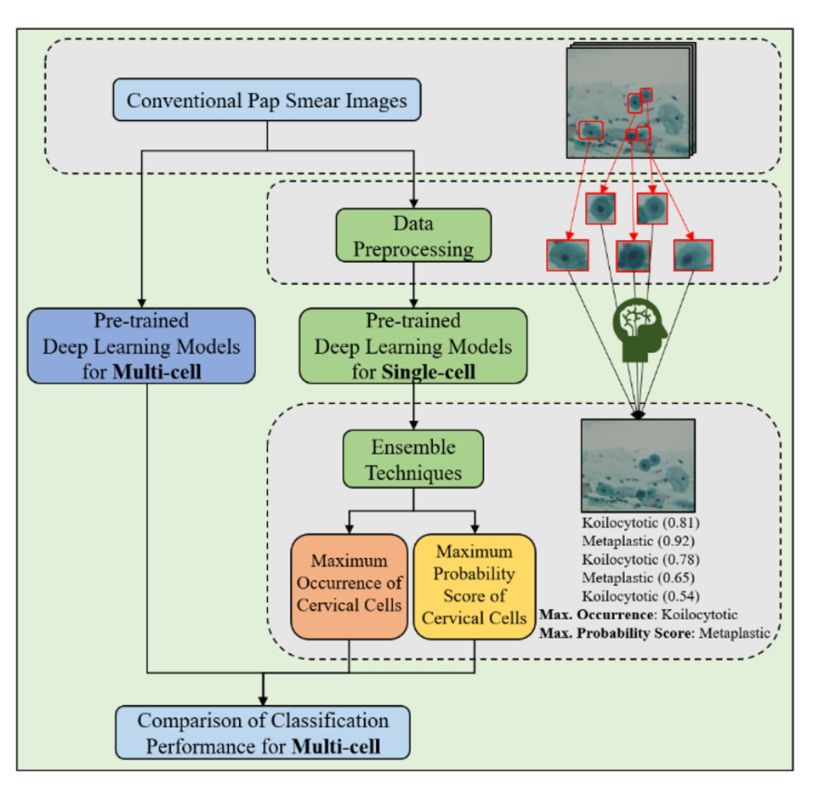
Cervical cancer screening allows the early signs of precancerous abnormalities in the cervix before they develop into invasive cancer. The Pap Smear is a widely used screening for early detection and prevention of cervical cancer. In many remote areas, the number of cytologists available to interpret pap smear screening tests is insufficient. This lack of personnel makes the test interpretation very time-consuming. To address this, deep learning techniques have been employed to detect cervical cancer cells and support cytologists. Therefore, an integrative approach with deep learning models and the ensemble techniques such as the maximum occurrence and the maximum probability score of cervical cells was proposed. The multi-cell assessment of the Pap smear slide allowed aggregate predictions of single cervical cell images using the proposed method. The classification results between pre-trained deep learning models and the proposed method were compared. In the experimental results, the proposed method can achieve an accuracy score of more than 97%, while the best pre-trained deep learning model can attain an accuracy score of more than 85%. Hence, the proposed method may have the potential to assist physicians or cytologists in the classification of cervical cell types for Pap Smear images.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
References
T. Hanprasertpong, K. Dhanaworavibul, K. Leetanaporn and J. Hanprasertpong, “Cervical Cancer Screening in Elderly Women,” Thai Journal of Obstetrics and Gynaecology, vol. 30, no. 6, pp. 370-375, 2022.
M. M. Rahaman et al., “DeepCervix: A deep learning-based framework for the classification of cervical cells using hybrid deep feature fusion techniques,” Computers in Biology and Medicine, vol. 136, p. 104649, 2021.
H. Sung et al., “Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries,” CA: A Cancer Journal for Clinicians, vol. 71, no. 3, pp. 209-249, 2021.
D. Saslow et al., “American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer,” CA: A Cancer Journal for Clinicians, vol. 62, no. 3, pp. 147-72, 2012.
E. Davey et al., “Effect of study design and quality on unsatisfactory rates, cytology classifications, and accuracy in liquid-based versus conventional cervical cytology: a systematic review,” The Lancet, vol. 367, no. 9505, pp. 122-32, 2006.
J. Rojanamatin et al., “Cancer in Thailand,” Bangkok Thailand: National Cancer Institute Ministry of Public Health, vol. X 2016-2018, 2021.
P. Phaliwong, P. Pariyawateekul, N. Khuakoonratt, W. Sirichai, K. Bhamarapravatana and K. Suwannarurk, “Cervical Cancer Detection between Conventional and Liquid Based Cervical Cytology: a 6-Year Experience in Northern Bangkok Thailand,” Asian Pacific Journal of Cancer Prevention, vol. 19, no. 5, pp. 1331-1336, 2018.
R. Mehrotra, S. Mishra, M. Singh and M. Singh, “The efficacy of oral brush biopsy with computer-assisted analysis in identifying precancerous and cancerous lesions,” Head & Neck Oncology, vol. 3, no. 1, p. 39, 2011.
A. Khamparia, D. Gupta, V. H. C. de Albuquerque, A. K. Sangaiah and R. H. Jhaveri, “Internet of health things-driven deep learning system for detection and classification of cervical cells using transfer learning,” The Journal of Supercomputing, vol. 76, no. 11, pp. 8590-8608, 2020.
V. Chandran et al., “Diagnosis of Cervical Cancer based on Ensemble Deep Learning Network using Colposcopy Images,” BioMed Research International, vol. 2021, p. 5584004, 2021.
H. Alquran et al., “Cervical Cancer Classification Using Combined Machine Learning and Deep Learning Approach,” Computers, Materials & Continua, vol. 72, no. 3, pp. 5117-5134, 2022.
M. Plissiti, P. Dimitrakopoulos, G. Sfikas, C. Nikou, O. Krikoni and A. Charchanti, “Sipakmed: A New Dataset for Feature and Image Based Classification of Normal and Pathological Cervical Cells in Pap Smear Images,” 2018 25th IEEE International Conference on Image Processing (ICIP), pp. 3144-3148, 2018.
J. Yosinski, J. Clune, Y. Bengio and H. Lipson, “How transferable are features in deep neural networks?,” Advances in Neural Information Processing Systems (NIPS), vol. 27, 2014.
S. J. Pan and Q. Yang, “A Survey on Transfer Learning,” IEEE Transactions on Knowledge and Data Engineering, vol. 22, no. 10, pp. 1345-1359, 2010.
K. Simonyan and A. Zisserman, “Very Deep Convolutional Networks for Large-Scale Image Recognition,” arXiv:1409.1556v6, 2014.
K. He, X. Zhang, S. Ren and J. Sun, “Identity Mappings in Deep Residual Networks,” Computer Vision – ECCV 2016, vol. 9908, pp. 630-645, 2016.
B. Zoph, V. Vasudevan, J. Shlens and Q. V. Le, “Learning Transferable Architectures for Scalable Image Recognition,” in 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 8697-8710, 2018.
M. T. a. Q. V. Le, “EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks,” CoRR, vol. abs/1905.11946, 2019. [Online]. Available: http://arxiv.org/abs/1905.11946.
R. R. Selvara ju, M. Cogswell, A. Das, R. Vedantam, D. Parikh and D. Batra, “GradCAM: Visual Explanations from Deep Networks via Gradient-Based Localization,” International Journal of Computer Vision, vol. 128, no. 2, pp. 336-359, 2020.
C. Duanggate, B. Uyyanonvara and T. Koanantakul, “A Review of Image Analysis and Pattern Classification Techniques for Automatic Pap Smear Screening Process,”International Conference on Embedded Systems and Intelligent Technology, 2008.
J. Jantzen and G. Dounias, “Analysis of Papsmear Image Data,” in Proceedings of the Nature-Inspired Smart Information Systems 2nd Annual Symposium, 2006.