Optimized CNN Model with Derived Kernels for Apnea Classification Application
Main Article Content
Abstract
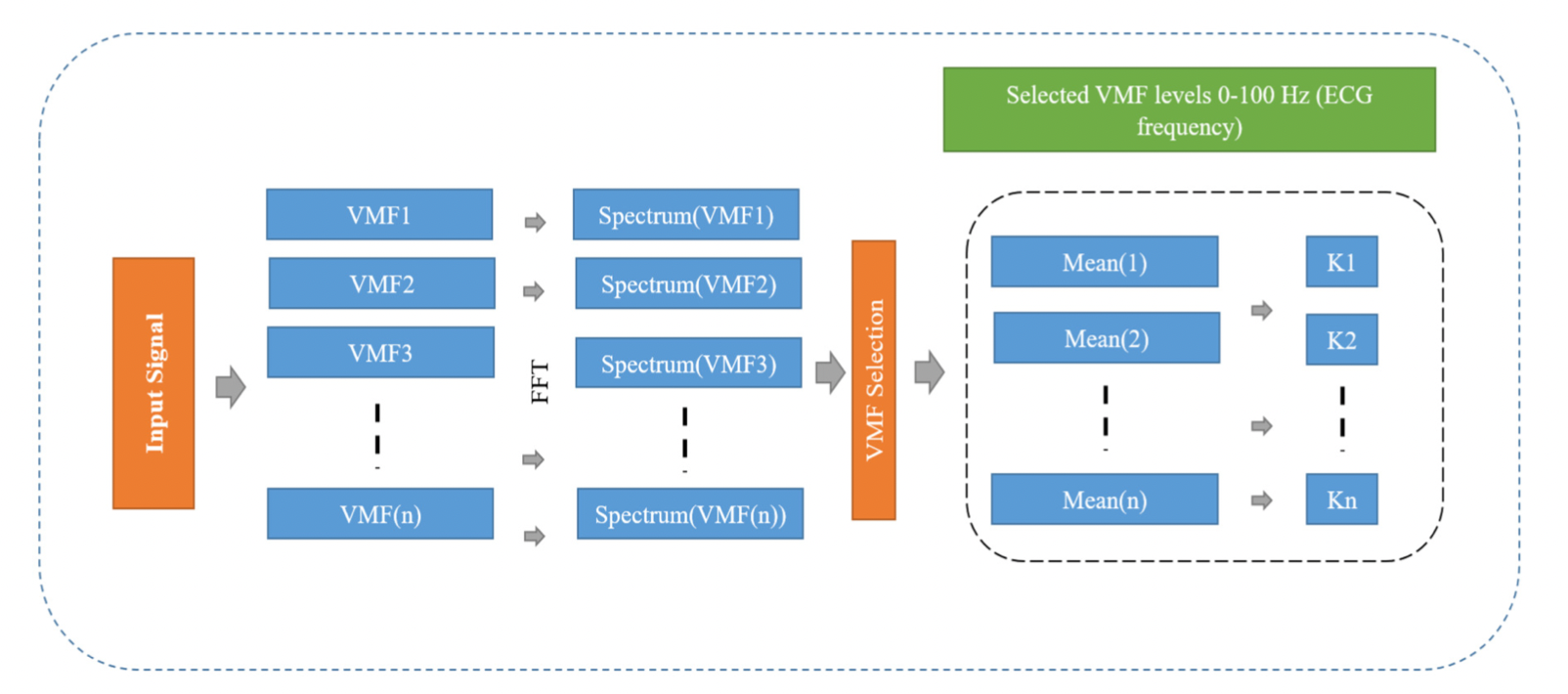
The sleep related disorders are common in both men and women, irrespective of their age. In recent years, there is an abrupt increase of interest in the prediction of sleep related disorders among the researchers. The most common data analysis approach utilize a large data set to predict the sleep apnea episode. The computational complexity of such predictions is high and most of the techniques are failing to select the optimal features. The recent trends show that the convolution neural network gaining the popularity because of its ability to select the optimal features and the dimension reduction property. On the other way, it is also important to address the noise issues in the acquired physiological data. The Variational Mode Decomposition (VMD) is an adaptive way of decomposing the different frequency levels, such that the noise level can be easily separated into the corresponding levels. This paper presents a novel technique for the optimal feature extraction from the Variational Mode Function (VMF) levels by using the concept of the Convolution Neural Network (CNN).
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
References
Koley, Bijoy Laxmi, and Debabrata Dey., “Real-Time Adaptive Apnea and Hypopnea Event Detection Methodology for Portable Sleep Apnea Monitoring Devices.”, IEEE transactions on biomedical engineering Vol.60, No.12, pp.3354– 3363, 2013.
Ciołek, Marcin., Maciej Niedźwiecki., Stefan Sieklicki., Jacek Drozdowski., and Janusz Siebert., “Automated Detection of Sleep Apnea and Hypopnea Events Based on Robust Airflow Envelope Tracking in the Presence of Breathing Artifacts”, IEEE journal of biomedical and health informatics, Vol.19, No.2, pp.418–429, 2014.
Jin, J. and Sánchez-Sinencio, E., “A home sleep apnea screening device with time-domain signal processing and autonomous scoring capability”, IEEE transactions on biomedical circuits and systems, Vol.9, No.1, pp.96–104, 2014.
Gutiérrez-Tobal., Gonzalo C., Daniel Alvarez., Félix Del Campo., and Roberto Hornero., “Utility of AdaBoost to detect sleep apnea-hypopnea syndrome from single-channel airflow”, IEEE transactions on biomedical engineering, Vol.63, No.3, pp.636– 646, 2015.
Karmakar., Chandan., Ahsan Khandoker., Thomas Penzel., Christoph Schöbel., and Marimuthu Palaniswami., “Detection of respiratory arousals using photoplethysmography (PPG) signal in sleep apnea patients”, IEEE journal of biomedical and health informatics, Vol.18, No.3, pp.1065–1073, 2013.
Domingues, A., Paiva, T. and Sanches, J.M., “Sleep and wakefulness state detection in nocturnal actigraphy based on movement information”, IEEE transactions on biomedical engineering, Vol.61, No.2, pp.426–434, 2013.
Mora., Guillermina Guerrero., Juha M., Kortelainen., Elvia Ruth Palacios Hernández., Mirja Tenhunen., Anna Maria Bianchi., and Martín O. Méndez., “Evaluation of pressure bed sensor for automatic SAHS screening”, IEEE transactions on instrumentation and measurement, Vol.64, No.7, pp.1935–1943, 2014.
Ahmad, S., Batkin, I., Kelly, O., Dajani, H.R., Bolic, M. and Groza, V., “Multiparameter physiological analysis in obstructive sleep apnea simulated with Mueller maneuver”, IEEE transactions on instrumentation and measurement, Vol.62, No.10, pp.2751–2762, 2013.
Hwang, S.H., Lee, H.J., Yoon, H.N., Lee, Y.J.G., Lee, Y.J., Jeong, D.U. and Park, K.S., “Unconstrained sleep apnea monitoring using polyvinylidene fluoride film-based sensor”, IEEE transactions on biomedical engineering, Vo.61, No.7, pp.2125–2134, 2014.
Kumar, T. S. and Kanhangad, V., “Gabor filter-based one-dimensional local phase descriptors for obstructive sleep apnea detection using single-lead ECG”, IEEE sensors letters, Vol. 2, No. 1, pp.1–4, 2018.
Zarei, A. and Asl, B. M., “Automatic detection of obstructive sleep apnea using wavelet transform and entropy-based features from single lead ECG signal”, IEEE journal of biomedical and health informatics, Vol. 23, No. 3, pp.1011–1021, 018.
Korkalainen, H., Aakko, J., Nikkonen, S., Kainulainen, S., Leino, A., Duce, B., Afara, I.O., Myllymaa, S., Töyräs, J. and Leppänen, T., “Accurate deep learning-based sleep staging in a clinical population with suspected obstructive sleep apnea”, IEEE journal of biomedical and health informatics, Vol. 24, No. 7, pp.2073–2081, 2019.
Sun, C., Fan, J., Chen, C., Li, W., and Chen, W., “A two-stage neural network for sleep stage classification based on feature learning, sequence learning, and data augmentation”, IEEE Access, Vol. 7, pp.109386–109397, 2019.
Gumery, P.-Y., Roux-Buisson, H., Meignen, S., Comyn, F. L., Dematteis, M., Wuyam, B., Pépin, J. L., and Lévy, P., “An adaptive detector of genioglossus EMG reflex using berkner transform for time latency measurement in OSA pathophysiological studies”, IEEE transactions on biomedical engineering, Vol. 52, No. 8, pp.1382– 1389, 2005.
Gutiérrez-Tobal., Gonzalo C., Daniel Álvarez., Andrea Crespo., Felix Del Campo., and Roberto Hornero., “Evaluation of machine-learning approaches to estimate sleep apnea severity from at-home oximetry recordings”, IEEE journal of biomedical and health informatics, Vol. 23, No. 2, pp.882–892, 2018.
Tran, V. P. and Al-Jumaily, A. A., “A novel oxygen-hemoglobin model for non-contact sleep monitoring of oxygen saturation”, IEEE Sensors Journal, Vol. 9, No. 24, pp.12325–12332, 2019. [17] Dragomiretskiy, K. and Zosso, D., “Variational mode decomposition”, IEEE transactions on signal processing, Vol. 62, No. 3,pp.531–544, 2013.
K. Dragomiretskiy and D.Zosso, “Variational mode decomposition,” IEEE transactions on sig- nal processing, vol. 62, No. 3,pp.531–544, 2013.
Dragomiretskiy, K. and Zosso, D., “Twodimensional variational mode decomposition”, International Workshop on Energy Minimization Methods in Computer Vision and Pattern Recognition, pp.197–208, 2015.
Salamon, J. and Bello, J.P., “Deep convolutional neural networks and data augmentation for environmental sound classification”, IEEE signal processing letters, Vol.24, No.3,pp.279–283, 2017.
Kalchbrenner, N., Grefenstette, E. and Blunsom, P., “A convolutional neural network for modelling sentences”, arXiv preprint arXiv:1404.2188, 2014.
Chen, Y., Jiang, H., Li, C., Jia, X. and Ghamisi, P., “Deep feature extraction and classification of hyperspectral images based on convolutional neural networks”, IEEE transactions on geoscience and remote sensing, Vol.54, No.10, pp.6232–6251, 2016.
Van Grinsven, B., Hoyng, C.B., Theelen, T.and Sánchez, C.I., “Fast convolutional neural network training using selective data sampling: Application to hemorrhage detection in color fundus images”, IEEE transactions on medical imaging, Vol.35, No.5, pp.1273–1284, 2016.
Lekha, S. and Suchetha, M., “Real-Time Non-Invasive Detection and Classification of Diabetes using Modified Convolution Neural Network”, IEEE journal of biomedical and health informatics, Vol.22, No.5, pp.1630–1636, 2017.
Lekha, S. and Suchetha, M., “A novel 1-D convolution neural network with SVM architecture for real-time detection applications”, IEEE Sensors Journal, Vol. 18, No. 2, pp.724–731, 2017.
De Falco, I., De Pietro, G., Della Cioppa, A., Sannino, G., Scafuri, U., and Tarantino, E., “Evolution-based configuration optimization of a deep neural network for the classification of obstructive sleep apnea episodes”, Future Generation Computer Systems, Vol. 98, pp.377–391, 2019.
Chen, K., Zhang, C., Ma, J., Wang, G., and Zhang, J., “Sleep staging from single-channel ECG with multi-scale feature and contextual information”, Sleep and Breathing, Vol. 23, No. 4, pp.1159–1167, 2019.
Wang, F.-T., Hsu, M.-H., Fang, S.-C., Chuang, L.- L., and Chan, H.-L., “The respiratory fluctuation index: A global metric of nasal airflow or thoracoabdominal wall movement time series to diagnose obstructive sleep apnea”, Biomedical Signal Processing and Control, Vol. 49, pp.250–262, 2019.
Viswabhargav, C. S., Tripathy, R., and Acharya, U. R., “Automated detection of sleep apnea using sparse residual entropy features with various dictionaries extracted from heart rate and EDR signals”, Computers in biology and medicine, Vol. 108, pp.20–30, 2019.
Banluesombatkul, N., Rakthanmanon, T., and Wilaiprasitporn, T., “Single channel ECG for obstructive sleep apnea severity detection using a deep learning approach.”, In TENCON 2018-2018 IEEE Region 10 Conference,pp.2011–2016, 2018.
Yang, T., Yu, L., Jin, Q., Wu, L. and He, B., “Localization of Origins of Premature Ventricular Contraction by Means of Convolutional Neural Network from 12-lead ECG”, IEEE transactions on biomedical engineering, Vol.65, No.7, pp.1662–1671, 2017.
Yu, L., Guo, Y., Wang, Y., Yu, J. and Chen, P., “Segmentation of fetal left ventricle in echocardiographic sequences based on dynamic convolutional neural networks”, IEEE transactions on biomedical engineering, Vol.64, No.8, pp.1886–1895, 2016.
Ince, T., Kiranyaz, S., Eren, L., Askar, M. and Gabbouj, M., “Real-time motor fault detection by 1-D convolutional neural networks”, IEEE transactions on industrial electronics, Vol.63, No.11, pp.7067–7075, 2016.
Kiranyaz, S., Ince, T. and Gabbouj, M., “Realtime patient-specific ECG classification by 1-D convolutional neural networks”, IEEE transactions on biomedical engineering, Vol.63, No.3, pp.664–675, 2015.
Vijayakumar, V., Case, M., Shirinpour, S. and He, B., “Quantifying and Characterizing Tonic Thermal Pain across Subjects from EEG Data using Random Forest Models”, IEEE transactions on biomedical engineering, Vol.64, No.12, pp.2988–2996, 2017.