Similarity in Sequences of COVID-19 Genetic Codes over Galois Field
Main Article Content
Abstract
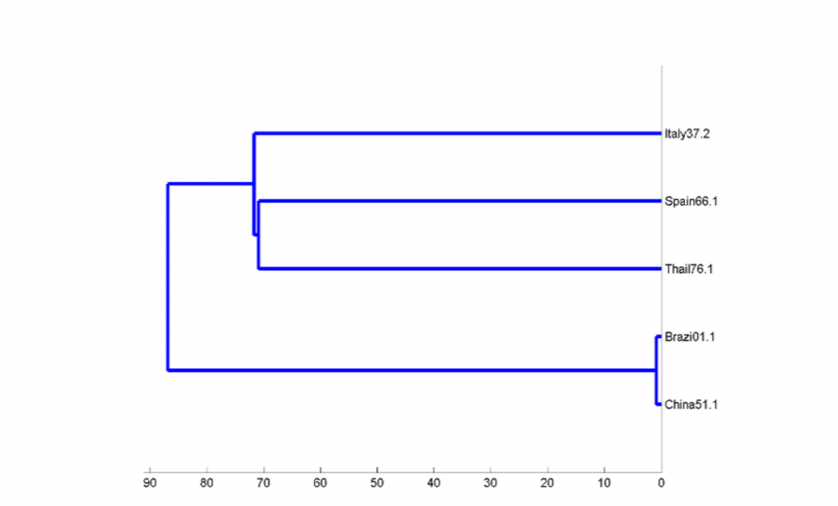
Coronavirus disease 2019 (COVID-19) outbreak is causing the economic and health problems for all countries. The origin based on genetic codes of its spreading is a significant key for identification and solution of the outbreak. The purpose of this research is to study the relations based on similarity measurement over Galois field amongst genetic codes of COVID-19. Galois field is an abstract structure for converting genetic codes to binary codes derived from polynomials and then similarity is measured from binary codes. The application is the investigation of the relations amongst the sequences of genetic codes of COVID-19 particles contaminated from waste water in Brazil, Spain, Italy and the sequences of COVID genetic codes in Thailand and China over Galois field. In addition, the relations amongst the sequences genetic COVID-19 codes from Wuhan markets, SARS and bat coronavirus genetic code are also investigated over Galois field. The results showed that the sequence of COVID-19 genetic codes in Brazil is possibly significant and related to the sequence in China, and vice versa, as well as the sequence of COVID-19 genetic codes at Wuhan market- LR757995.1 is possibly transmitted from Bat coronavirus RaTG13 genetic code to human in China.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
References
Z. Wu et al., “Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases,” ISME Journal, Vol. 10, No.3, pp. 609–620, 2016.
P. Zhou et al., “A pneumonia outbreak associated with a new coronavirus of probable bat origin,” Nature, Vol. 579, pp. 270–273, 2020.
F. Gislaine et al., “SARS-CoV-2 in human sewage in Santa Catalina, Brazil,” medRxiv, November 2019, 2020.06.26.20140731, 2019.
Z. Lara, and M. M. Marcelo, “The COVID-19 pandemic in Brazil: An urge for coordinated public health policies,” ⟨hal-02881690v2⟩, 2020.
C. M. Gemma et al., “Sentinel surveillance of SARS-CoV-2 in wastewater anticipates the occurrence of COVID-19 cases,” medRxiv, 2020.06.13.20129627, 2020.
L. R. Giuseppina et al., “First detection of SARS-CoV-2 in untreated waste waters in Italy,” Science of the Total Environment, Vol. 736, pp.139652, 2020.
R. Sunthornwat, E. J. Moore, and Y. Temtanapat, “Detecting and Classifying Mutations in Genetic Code with an Application to Beta-Thalassemia,” Science Asia, Vol. 37, No. 1, pp. 51-61, 2011.
P. Kirdwichai and M. Baksh, “The analysis of genomewide SNP data using nonparametric and kernel machine regression,” The Journal of Applied Science, Vol. 18, pp. 20-30, 2019.
J. M. Kim et al., “Identification of Coronavirus Isolated from a Patient in Korea with COVID-19,” Osong Public Health and Research Perspectives, Vol. 11, pp. 3-7, 2020.
J. C. Perez, “Wuhan COVID-19 Synthetic Origins and Evolution,” International Journal of Research – GRANTHAALAYAH, Vol. 8, pp. 285-324, 2020.
J. C. Perez and L. Montagnier, “COVID-19, SARS and Bats Coronaviruses Genomes Unexpected Exogeneous RNA Sequences,” OSF Preprints, 2020.
Z. Z. Yong and C. H. Edward, “A Genomic Perspective on the Origin and Emergence of SARS-CoV-2,” Cell, Vol. 181, pp. 223-227, 2020.
B. Robson, “Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus,” Computers in Biology and Medicine, Vol.119, pp. 103670, 2020.
D. Ellinghaus et al. “Genomewide Association Study of Severe Covid-19 with Respiratory Failure,” New England Journal of Medicine, Vol. 383, pp. 1522-1534, 2020.
X. C. Tang et al., “Prevalence and genetic diversity of coronaviruses in bats from China,” Journal of Virology, Vol. 80, pp. 7481-7490, 2006.
S. Robert, M. Eberto and G. Ricardo, “A Genetic code Boolean structure I. The meaning of Boolean deductions,” Journal of Mathematical Biology, Vol. 67, pp. 1-14, 2005.
A. G. Joseph, Contemporary abstract algebra, 4th ed, Houghton Mifflin, Boston, 1998.
E. Savas and C. Koc, “Finite Field Arithmetic for Cryptography,” Circuits and Systems Magazine- IEEE, Vol. 10, pp. 40 - 56, 2010.
D. J. Witherspoon et al., “Genetic Similarities Within and Between Human Populations,” Genetics, Vol. 176, pp. 351–359, 2007.
K. Jitobaom et al., “Codon usage similarity between viral and some host genes suggests a codon-specific translational regulation,” Heliyon, Vol. 6, pp. e03915, 2020.
P. S. Eldra, R. B. Linda and W. M. Diana, Biology, 6th ed, Brooks/Cole, Australia, 2002.
R. Lewis, Human Genetics: Concepts and Applications, 6th ed, McGraw-Hill, Boston, 2005.
J. Han, M. Kamber and J. Pei, Data mining: Concepts and techniques, 3rd ed, Morgan Kaufmann Publishers, Waltham Mass, 2012.
W. K. Härdle and H. Zdenek. Multivariate statistics exercises and solutions, London, Springer, 2015.
W. K. Härdle and L. Simar, Applied multivariate statistical analysis, 4th ed, Springer, London, 2019.
National Centre of Biotechnology Information, “NCBI SARS-CoV-2 Resources,” 2020. [online] Available at: https://www.ncbi.nlm.nih.gov/sars-cov-2
N. Kaur et al., “Genetic comparison among various coronavirus strains for the identification of potential vaccine targets of SARS-CoV2,” Infection, Genetics and Evolution, Vol. 89, pp. 1-15, 2021.
L. Vijgen et al., “Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event,” Journal of Virology, Vol. 79, pp. 1595-1604, 2005.
M. R. Islam et al., “Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity,” Scientific Reports, Vol. 10, pp. 1-9, 2020.
Y. A. Helmy et al., “The COVID-19 Pandemic: A Comprehensive Review of Taxonomy, Genetics, Epidemiology, Diagnosis, Treatment, and Control,” Journal of Clinical Medicine, Vol. 9, pp. 1-29, 2020.