Discriminative Image Enhancement for Robust Cascaded Segmentation of CT Images
Main Article Content
Abstract
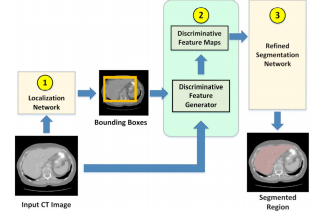
Objective: Cascaded/attention-based neural network has become common in image segmentation. This work proposes to improve its robustness by adding discriminative image enhancement to its attention mechanism. Unlike prior work, this image enhancement can also be applied as data augmentation and easily adapted for existing models. Its generalization can improve accuracy across multiple segmentation tasks and datasets. Methods: The method first localizes a target organ in a 2D fashion to obtain a tight neighborhood of the organ in each slice. Next, the method computes an HU histogram of a region combined from multiple 2D neighborhoods. This allows the method to adaptively handle HU-range difference among images. Then, HUs are nonlinearly stretched through a parameterized mapping function providing discriminative features for neural network. Varying the function parameters creates different intensity distribution of the target region. This effectively enhances and augments image data at the same time. The HU-reassigned region is then fed to a segmentation model for training. Results: Our experiments on liver and kidney segmentation showed that even a simple cascaded 2D U-Net model could deliver competitive performance in a variety of datasets. In addition, cross-validation and ablation analysis indicated robustness of the method even when the number of original training samples was limited. Conclusion: With the proposed technique, a simple model with limited training data can deliver competitive performance. Significance: The method significantly improves robustness of a trained model and is ready for generalization to other segmentation tasks and attention-based models. Accurate models can be simpler to save computing resources.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
References
H. Ryu et al., Total kidney and liver volume is a major risk factor for malnutrition in ambulatory patients with autosomal dominant polycystic kid ney disease, BMC Nephrol., vol. 18, no. 1, p. 22, Dec. 2017.
A. L. Simpson et al., A large annotated medical image dataset for the development and evaluation of segmentation algorithms, in arxiv.org, 2019.
X. Li, H. Chen, X. Qi, Q. Dou, C.-W. Fu, and P.-A. Heng, H-DenseUNet: Hybrid Densely Con nected UNet for Liver and Tumor Segmentation From CT Volumes, IEEE Trans. Med. Imaging, vol. 37, no. 12, pp. 2663 2674, Dec. 2018.
F. Isensee and K. H. Maier-Hein, An attempt at beating the 3D U-Net, in arxiv.org, 2019.
T. Y. Lin, P. Goyal, R. Girshick, K. He, and P. Dollar, Focal Loss for Dense Object Detection, in Proceedings of the IEEE International Confer ence on Computer Vision, 2017, vol. 2017-Octob, pp. 2999 3007.
F. B.V., zyr/keras-retinanet: Keras imple mentation of RetinaNet object detection, [Online]. Available: github.com/fizyr/ keras-retinanet. [Accessed: 23-Nov-2019].
P. Bilic et al., The Liver Tumor Segmentation Benchmark (LiTS), in arxiv.org, 2019.
L. Soler et al., 3D Image reconstruction for comparison of algorithm database : A patient speci c anatomical and medical im age database, [Online]. Available: ircad.fr/ softwares/3dircadb.
B. Van Ginneken, T. Heimann, and M. Styner, 3D segmentation in the clinic: A grand chal lenge, in In: MICCAI Workshop on 3D Segmen tation in the Clinic: A Grand Challenge (2007), 2007.
P. F. Jaeger et al., Retina U-Net: Embarrass ingly Simple Exploitation of Segmentation Super vision for Medical Object Detection, Nov. 2018.
A. Suzani, A. Seitel, Y. Liu, S. Fels, R. N. Rohling, and P. Abolmaesumi, Fast automatic vertebrae detection and localization in pathologi cal CT scans - a deep learning approach, in Lec ture Notes in Computer Science, 2015, vol. 9351, pp. 678 686.
R. Janssens, G. Zeng, and G. Zheng, Fully auto matic segmentation of lumbar vertebrae from CT images using cascaded 3D fully convolutional net works, in 2018 IEEE 15th Int. Symp. on Biomed ical Imaging (ISBI 2018), 2018, pp. 893 897.
I. I. N. Lessmann, B. van Ginneken, P. A. de Jong, Iterative convolutional neural networks for automatic vertebra identi cation and segmenta tion in CT images, in Medical Imaging 2018: Im age Processing, 2018, vol. 10574, p. 7.
H. Jiang, T. Shi, Z. Bai, and L. Huang, AHC Net: An Application of Attention Mechanism and
Hybrid Connection for Liver Tumor Segmenta tion in CT Volumes, IEEE Access, vol. 7, pp. 24898 24909, 2019.
A. Myronenko and A. Hatamizadeh, 3D Kid neys and Kidney Tumor Semantic Segmentation using Boundary-Aware Networks, in arxiv.org, 2019.
Y. Yuan, Hierarchical Convolutional Deconvolutional Neural Networks for Automatic Liver and Tumor Segmentation, Oct. 2017.
N. Heller et al., The KiTS19 Challenge Data: 300 Kidney Tumor Cases with Clinical Context, CT Semantic Segmentations, and Surgical Out comes.
S. Zheng, B. Fang, L. Li, M. Gao, and Y. Wang, A variational approach to liver segmentation us ing statistics from multiple sources, Phys. Med. Biol., vol. 63, no. 2, Jan. 2018.
W. Wu, Z. Zhou, S. Wu, and Y. Zhang, Auto matic Liver Segmentation on Volumetric CT Im ages Using Supervoxel-Based Graph Cuts, Com put. Math. Methods Med., vol. 2016, pp. 1 14, 2016.
D. Keshwani, Y. Kitamura, and Y. Li, Compu tation of total kidney volume from CT images in autosomal dominant polycystic kidney disease us ing multi-task 3D convolutional neural networks, in Lecture Notes in Computer Science, 2018, vol. 11046 LNCS, pp. 380 388.
F. Isensee et al., nnU-Net: Self-adapting Frame work for U-Net-Based Medical Image Segmenta tion, in Informatik aktuell, 2019, p. 22.
M. Perslev, E. Dam, A. Pai, and C. Igel, One Network To Segment Them All: A General, Lightweight System for Accurate 3D Medical Im age Segmentation, Med. Image Comput. Com put. Assist. Interv., vol. 11, no. 2, p. 24001, 2019.
C. Huang, H. Han, Q. Yao, S. Zhu, and S. K. Zhou, 3D U-Net: A 3D Universal U-Net for Multi-domain Medical Image Segmentation, 2019, pp. 291 299.
Y. Qin et al., Autofocus layer for semantic seg mentation, in Lecture Notes in Computer Sci ence, 2018, vol. 11072 LNCS, pp. 603 611.
B. Kayalibay, G. Jensen, and P. van der Smagt, CNN-based Segmentation of Medical Imaging Data, Jan. 2017.
P. F. Christ et al., Automatic Liver and Tu mor Segmentation of CT and MRI Volumes using Cascaded Fully Convolutional Neural Networks, in arxiv.org, 2017.
O. Ronneberger, P. Fischer, and T. Brox, U Net: Convolutional Networks for Biomedical Im age Segmentation, Springer, Cham, 2015, pp. 234 241.
K. Yin et al., Deep learning segmentation of kid neys with renal cell carcinoma., J. Clin. Oncol.,
vol. 37, no. 15_suppl, pp. e16098 e16098, May 2019.
J. A. O'Reilly, M. Sangworasil, and T. Matsuura, Kidney and Kidney Tumor Segmentation using a Logical Ensemble of U-nets with Volumetric Val idation, arxiv.org, Aug. 2019.
K. Xia, H. Yin, P. Qian, Y. Jiang, and S. Wang, Liver Semantic Segmentation Algorithm Based on Improved Deep Adversarial Networks in Combination of Weighted Loss Function on Ab dominal CT Images, IEEE Access, vol. 7, pp. 96349 96358, Jul. 2019.
L. C. Chen, Y. Zhu, G. Papandreou, F. Schro , and H. Adam, Encoder-decoder with atrous sep arable convolution for semantic image segmenta tion, Lect. Notes Comput. Sci., vol. 11211 LNCS, pp. 833 851, Feb. 2018.
L. C. Chen, G. Papandreou, I. Kokkinos, K. Murphy, and A. L. Yuille, DeepLab: Seman tic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs, IEEE Trans. Pattern Anal. Mach. Intell., vol. 40, no. 4, pp. 834 848, 2018.
L. Wang, R. Chen, S. Wang, N. Zeng, X. Huang, and C. Liu, Nested Dilation Network (NDN) for Multi-Task Medical Image Segmentation, IEEE Access, vol. 7, pp. 44676 44685, 2019.
R. Lamba et al., CT Houns eld numbers of soft tissues on unenhanced abdominal CT scans: Variability between two di erent manufacturers' MDCT scanners, Am. J. Roentgenol., vol. 203, no. 5, pp. 1013 1020, Nov. 2014.
O. Ronneberger, P. Fischer, and T. Brox, U net: Convolutional networks for biomedical im age segmentation, in Lecture Notes in Computer Science, vol. 9351, Springer, Cham, 2015, pp. 234 241.
N. Srivastava, G. Hinton, A. Krizhevsky, I. Sutskever, and R. Salakhutdinov, Dropout: A Simple Way to Prevent Neural Networks from Over tting, J. Mach. Learn. Res., vol. 15, pp. 1929 1958, 2014.
S. Io e and C. Szegedy, Batch Normalization: Accelerating Deep Network Training by Reduc ing Internal Covariate Shift, in Proceedings of the 32nd Int. Conf. on Machine Learning, 2015, vol. 37, pp. 448 456.
A. A. Taha and A. Hanbury, Metrics for evalu ating 3D medical image segmentation: Analysis, selection, and tool, BMC Med. Imaging, vol. 15, no. 1, Aug. 2015.
K. Sharma et al., Automatic Segmentation of Kidneys using Deep Learning for Total Kidney Volume Quanti cation in Autosomal Dominant Polycystic Kidney Disease, Sci. Rep., vol. 7, no. 1, Dec. 2017.
G. Li, X. Chen, F. Shi, W. Zhu, J. Tian, and D. Xiang, Automatic Liver Segmentation Based
on Shape Constraints and Deformable Graph Cut in CT Images, IEEE Trans. Image Process., vol. 24, no. 12, pp. 5315 5329, Dec. 2015.
X. Lu, Q. Xie, Y. Zha, and D. Wang, Fully automatic liver segmentation combining multi dimensional graph cut with shape information in 3D CT images, Sci. Rep., vol. 8, no. 1, p. 10700, Dec. 2018.
F. Lu, F. Wu, P. Hu, Z. Peng, and D. Kong, Au tomatic 3D liver location and segmentation via convolutional neural network and graph cut, Int. J. Comput. Assist. Radiol. Surg., vol. 12, no. 2, pp. 171 182, Feb. 2017.
KiTS19 Results. [Online]. Available: http: //results.kits-challenge.org/miccai2019/. [Accessed: 22-Nov-2019].
Y. Zheng et al., Automatic liver segmentation based on appearance and context information, Biomed. Eng. Online, vol. 16, no. 1, p. 16, Dec. 2017.