Hybrid Learning of Vessel Segmentation in Retinal Images
Main Article Content
Abstract
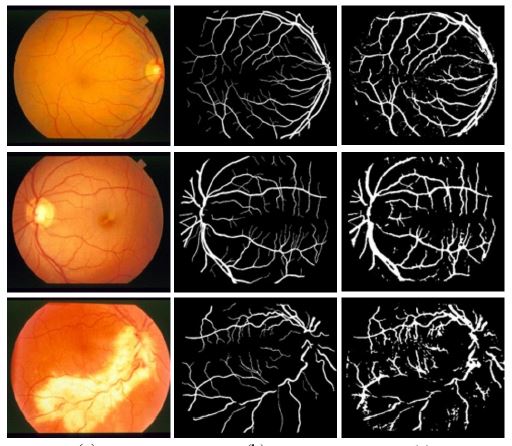
This paper aims to develop a technique of vessel segmentation in retinal images. Interpreting the segmented vessels is necessary for the automatic detection of the severe stage of the diabetic retinopathy. Thus, it is important to have the technique for segmenting vessels in an automatic way with high performance, for the sake of further analysis. In this paper, the proposed method is developed based on the double layer combining supervised and non-supervised learning aspects. The first layer is to detect the initial seeds of vessels using the supervised learning. It learns based on three types of features including green intensity, line operators, and Gabor filters. Then, the support vector machine (SVM) is applied as the classification tool. In the second layer, the segmentation results from the first layer is further revised and completed using the non-supervised learning. The morphological operations with the watershed technique are applied on the results obtained from the first layer, to remain with the segmented pixels with high confidential to be vessels. Then, these pixels are used as the initial seeds of foreground in the iterative graph cut. As the result, the more completed and comprehensive foreground (i.e. vessels) can be obtained. The proposed method is evaluated using two well-known datasets including DRIVE and STARE. The experimental results show the promising performance of the proposed method when compared with other existing methods in the literature.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
References
A. Imran, J. Li, Y. Pei, J.-J. Yang, and Q. Wang, “Comparative analysis of vessel segmentation techniques in retinal images," IEEE Access, vol. 7, pp. 114862-114887, 2019.
J. Staal, M. D. Abr_amo_, M. Niemeijer, M. A. Viergever, and B. Van Ginneken, “Ridge-based vessel segmentation in color images of the retina," IEEE transactions on medical imaging, vol. 23, no. 4, pp. 501-509, 2004.
J. Zhang, Y. Cui, W. Jiang, and L. Wang, “Blood vessel segmentation of retinal images based on neural network," in International Conference on Image and Graphics. Springer, 2015, pp. 11-17.
D. Paulus, S. Chastel, and T. Feldmann, “Vessel segmentation in retinal images," in Medical Imaging 2005: Physiology, Function, and Structure from Medical Images, vol. 5746. International Society for Optics and Photonics, 2005, pp. 696-706.
A. G. Salazar-Gonzalez, D. Kaba, Y. Li, and X. Liu, “Segmentation of the blood vessels and optic disk in retinal images." IEEE J. Biomedical and Health Informatics, vol. 18, no. 6, pp. 1874-1886, 2014.
E. Ricci and R. Perfetti, Retinal blood vessel segmentation using line operators and support vector classification," IEEE transactions on medical imaging, vol. 26, no. 10, pp. 1357-1365, 2007.
R. Kharghanian and A. Ahmadyfard, “Retinal blood vessel segmentation using gabor wavelet and line operator," International Journal of Machine Learning and Computing, vol. 2, no. 5, p. 593, 2012.
R. Perfetti, E. Ricci, D. Casali, and G. Costantini, “Cellular neural networks with virtual template expansion for retinal vessel segmentation," IEEE Transactions on Circuits and Systems II: Express Briefs, vol. 54, no. 2, pp. 141-145, 2007.
Z. Han, Y. Yin, X. Meng, G. Yang, and X. Yan, “Blood vessel segmentation in pathological retinal image," in Data Mining Workshop (ICDMW), 2014 IEEE International Conference on. IEEE, 2014, pp. 960-967.
D. S. S. Raja, S. Vasuki, and D. R. Kumar, “Performance analysis of retinal image blood vessel segmentation," Advanced Computing, vol. 5, no.
/3, p. 17, 2014.
D. Marin, A. Aquino, M. E. Gegundez-Arias, and J. M. Bravo, “A new supervised method for blood vessel segmentation in retinal images by using gray-level and moment invariants-based features," IEEE transactions on medical imaging, vol. 30, no. 1, p. 146, 2011.
Q. Jin, Z. Meng, T. D. Pham, Q. Chen, L. Wei, and R. Su, “Dunet: A deformable network for retinal vessel segmentation," Knowledge-Based
Systems, vol. 178, pp. 149-162, 2019.
D. Yang, M. Ren, and B. Xu, “Retinal blood vessel segmentation with improved convolutional neural networks," Journal of Medical Imaging and Health Informatics, vol. 9, no. 6, pp. 1112-1118, 2019.
S. Guo, K. Wang, H. Kang, Y. Zhang, Y. Gao, and T. Li, “Bts-dsn: Deeply supervised neural network with short connections for retinal vessel segmentation," International journal of medical informatics, vol. 126, pp. 105-113, 2019.
A. Hatamizadeh, H. Hosseini, Z. Liu, S. D. Schwartz, and D. Terzopoulos, “Deep dilated convolutional nets for the automatic segmentation of retinal vessels," arXiv preprint arXiv:1905.12120, 2019.
Z. Fan, J. Mo, and B. Qiu, “Accurate retinal vessel segmentation via octave convolution neural network," arXiv preprint arXiv:1906.12193, 2019.
K. J. Noh, S. J. Park, and S. Lee, “Scale-space approximated convolutional neural networks for retinal vessel segmentation," Computer methods and programs in biomedicine, vol. 178, pp. 237-246, 2019.
Q. Jin, Q. Chen, Z. Meng, B. Wang, and R. Su, “Construction of retinal vessel segmentation models based on convolutional neural network," Neural Processing Letters, pp. 1-18, 2019.
J. Li, “An empirical comparison between svms and anns for speech recognition," in The First Instructional Conf. on Machine Learning, vol. 951, 2003, p. 2003.
E. Reinhard, M. Adhikhmin, B. Gooch, and P. Shirley, “Color transfer between images," IEEE Computer graphics and applications, vol. 21, no. 5, pp. 34-41, 2001.
W. Kusakunniran, A. Wiratsudakul, U. Chuachan, S. Kanchanapreechakorn, and T. Imaromkul, Automatic cattle identification based on fusion of texture features extracted from muzzle images," in 2018 IEEE International Conference on Industrial Technology (ICIT). IEEE, 2018, pp. 1484-1489.
D. Barina, “Gabor wavelets in image processing," arXiv preprint arXiv:1602.03308, 2016.
M. M. Fraz, P. Remagnino, A. Hoppe, S. Velastin, B. Uyyanonvara, and S. Barman, “A supervised method for retinal blood vessel segmentation using line strength, multiscale gabor and morphological features," in Signal and Image Processing Applications (ICSIPA), 2011 IEEE International Conference on. IEEE, 2011, pp. 410-415.
I. Steinwart and A. Christmann, Support vector machines. Springer Science & Business Media, 2008.
A. Iranmehr, H. Masnadi-Shirazi, and N. Vasconcelos, “Cost-sensitive support vector machines," Neurocomputing, 2019.
W. Kusakunniran, K. Ngamaschariyakul, C. Chantaraviwat, K. Janvittayanuchit, and K. Thongkanchorn, “A thai license plate localization using svm," in 2014 International Computer Science and Engineering Conference
(ICSEC). IEEE, 2014, pp. 163-167.
P. Druzhkov, V. Erukhimov, N. Y. Zolotykh, E. Kozinov, V. Kustikova, I. Meerov, and A. Polovinkin, “New object detection features in the opencv library," Pattern Recognition and Image Analysis, vol. 21, no. 3, p. 384, 2011.
C.-C. Chang and C.-J. Lin, “Libsvm: A library for support vector machines," ACM transactions on intelligent systems and technology (TIST), vol. 2, no. 3, p. 27, 2011.
G. Bradski and A. Kaehler, Learning OpenCV: Computer vision with the OpenCV library. “O'Reilly Media, Inc.", 2008.
P. K. Ghosh and R. M. Haralick, “Mathematical morphological operations of boundary-represented geometric objects," Journal of Mathematical Imaging and Vision, vol. 6, no. 2-3, pp. 199-222, 1996.
J. B. Roerdink and A. Meijster, “The watershed transform: Definitions, algorithms and parallelization strategies," Fundamenta informaticae, vol. 41, no. 1, 2, pp. 187-228, 2000.
C. Rother, V. Kolmogorov, and A. Blake, “Grab-cut: Interactive foreground extraction using iterated graph cuts," in ACM transactions on graphics (TOG), vol. 23, no. 3. ACM, 2004, pp. 309-314.
M. Eapen, R. Korah, and G. Geetha, “Swarm intelligence integrated graph-cut for liver segmentation from 3d-ct volumes," The Scientific World Journal, vol. 2015, 2015.
L. Massoptier and S. Casciaro, “Fully automatic liver segmentation through graph-cut technique," in 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE, 2007, pp. 5243-5246.
J. Cha, M. M. Farhangi, N. Dunlap, and A. A. Amini, “Segmentation and tracking of lung nodules via graph-cuts incorporating shape prior and motion from 4d ct," Medical physics, vol. 45, no. 1, pp. 297-306, 2018.
J. won Cha, N. Dunlap, B. Wang, and A. Amini, “3d segmentation of lung ct data with graph-cuts: analysis of parameter sensitivities," in Medical Imaging 2016: Biomedical Applications in Molecular, Structural, and Functional Imaging, vol. 9788. International Society for Optics and Photonics, 2016, p. 97882O.
J. F. Talbot and X. Xu, “Implementing grab-cut," Brigham Young University, vol. 3, 2006.
T. Malmer, “Image segmentation using grab-cut," IEEE Transactions on Signal Processing, vol. 5, no. 1, pp. 1-7, 2010.
A. Blake, C. Rother, M. Brown, P. Perez, and P. Torr, “Interactive image segmentation using an adaptive gmmrf model," in European conference on computer vision. Springer, 2004, pp. 428-441.
Y. Boykov and V. Kolmogorov, “An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision," IEEE Transactions on Pattern Analysis & Machine Intelligence, no. 9, pp. 1124-1137, 2004.
M. Stoer and F. Wagner, “A simple min-cut algorithm," Journal of the ACM (JACM), vol. 44, no. 4, pp. 585-591, 1997.
A. Hoover and M. Goldbaum, “Locating the optic nerve in a retinal image using the fuzzy convergence of the blood vessels," IEEE transactions on medical imaging, vol. 22, no. 8, pp. 951-958, 2003.
C. Wang, Z. Zhao, Q. Ren, Y. Xu, and Y. Yu, “Dense u-net based on patch-based learning for retinal vessel segmentation," Entropy, vol. 21, no. 2, p. 168, 2019.
B. Zupan, E. T. Keravnou, and N. Lavrac, Intelligent Data Analysis in Medicine and Pharmacology. Kluwer Academic Publishers, 1997.